Courses in Practical DNA Microarray Analysis 2008
![]()
2008 May 5-8 in Munich
Registration is closed.
To get on the waiting-list, please send a short note that explains your existing skills and experience with (a) microarrays, (b) statistics, and (c) computer programming and your motivation for taking this course to t.beissbarth@dkfz.de.
If you bring your own laptop, be sure to have installed the following packages:
affy, Biobase, marray, multtest, limma, vsn, arrayMagic, cluster, RColorBrewer, hgu95av2, hgu95av2cdf, estrogen,
ALL, annotate, genefilter, multtest, ROC, isis, pamr, e1071, MCRestimate, GlobalAncova, globaltest, GOstats,
hgu133a, Rgraphviz.
Datasets and additional packages used in the practical sessions can be found in the data directory, links to the tutorial slides and practical exercises in the table below.
![]()
| Monday, May 05 - First Analysis steps | |||
| 09.00-09.30 | Introduction and overview | Ulrich Mansmann | |
| 09.30-12.30 | Quality control, normalization and design | Tim Beissbarth | |
| 14.00-17.00 | Exercises: Introduction to R, cDNA data, affy data | Mansmann, Hummel, Beissbarth | |
| Tuesday, May 06 - Exploratory analysis | |||
| 09.00-10.30 | Differential gene expression | Ulrich Mansmann | |
| 10.30-12.00 | Clustering | Adrian Alexa | |
| 13.30-17.00 | Exercises: Differential gene expression Clustering |
Mansmann, Hummel, Alexa | |
| Wednesday, May 07 - Molecular Diagnosis | |||
| 09.00-10.30 | Molecular Diagnosis | Holger Fröhlich | |
| 10.30-12.00 | Classification with PAM and Random Forest | Markus Ruschhaupt | |
| 13.30-17.00 | Exercises: Molecular Diagnosis | Ruschhaupt, Fröhlich, Alexa | |
| Thursday, May 08 - Pathways | |||
| 09.00-10.00 | Computational Inference of Cellular Networks | Achim Tresch | |
| 10.15-11.15 | Group testing: global tests, holistic approaches | Manuela Hummel | |
| 11.30-12.30 | Scoring Gene Ontology terms | Adrian Alexa | |
| 13.30-17.00 | Exercises: Geneset Enrichment, Global testing, topGO | Tresch, Hummel, Alexa | |
![]()
Background Knowledge
Ideally, you are interested in mathematical and statistical problems and are familiar with at least one programming language. This course focusses on the practical side of gene expression data analysis. However, data analysis without understanding the statistical background is in general impossible. We strongly recommend you to refresh your mathematical and programming skills before attending the course.
Please use the links to software and literature to prepare yourself before the course begins.
R and Bioconductor
In the afternoon exercises you will learn how to analyze data using the statistical computing environment R [http://www.r-project.org] and BioConductor [http://www.bioconductor.org], an open source software for bioinformatics. R sources and package sources can be downloaded from The Comprehensive R Archive Network at http://cran.r-project.org.
This is a course in microarray analysis -- not an introduction to R. Please read the Introduction to R before the course begins.
Bring your own data
You are encouraged to bring some of your own data to the course (e.g. genepix files or CEL/CDF). We will use this during the exercises. If you expect to have own data only later in the year, it may in fact be advantageous also to register for one of the later courses.
Participants
| Harmjan Kuipers | LMU Munich -IFI | Germany | |
| Winfried Hofmann | Medizinische Hochschule Hannover, OE 5120 | Germany | |
| Wenjing Zhou | Uppsala University | Sweden | |
| Aleksandra Swiercz | Poznan University of Technology, Inst. Comp.Sc. | Poland | |
| Anna Widelska | Poznan University of Technology,Inst.Comp.Scienc | Poland | |
| Sergey Ustinov | National University of Ireland, Galway | Ireland | |
| Inka Praulich | Medizinische Hochschule Hannover | Germany | |
| Yinyin Yuan | Warwick University | UK | |
| Josep Pareja Gomez | Pompeu Fabra University | Spain | |
| sriram kannan | individual trying for Phd | India | |
| Diana Santacruz | University of Saarland | Germany | |
| Andreas Mayer | gene center/LMU | 81377 Munich | Cancelled |
| Guvanch Ovezmuradov | Genetics Institute University of Cologne | Germany | |
| Michal Swierniak | Cancer Centre, Institute of Oncology | Poland | |
| Aleksandra Pfeifer | Cancer Centre, Institute of Oncology | Poland | |
| Wouter Van Gool | VIB (www.vib.be) | 9052 GENT, BELGIUM | |
| Nina Pälmke | Epigenetics, University of Saarland | Germany | |
| Stefano Torti | Max Planck Institute for Plant Breeding Research | Germany | |
| Stefan Franke | University Hospital, Dept. Ped. Oncology | Germany | |
| Michal Jarzab | MSC Memorial Cancer Center, Gliwice | Poland | |
| Malgorzata Oczko-Wojciechowska | MSC Memorial Cancer Center and Institute of Onco | Poland | |
| jeevitesh ms | IBAB | INDIA | |
| Christina Poehler | DKFZ | Germany | |
| Salim Lardjane | Helmholtz Center Munich, IBB | Germany | |
| Zlatana Pasalic | Helmholz Zentrum Munich | Germany | |
| Marko Petek | National Institute of Biology | Slovenia | |
| Jana Silhava | Faculty of Information Technology, BUT | Czech Republic | |
| Kim Van Deun | Ghent University | Belgium | Cancelled |
| Vindi Jurinovic | LMU - IBE | Germany | |
| Zheng Yu | TU Muenchen | Germany | |
| Romain Fenouil | CIML centre CNRS INSERM | France | |
| Karine Sá Ferreira | University of Freiburg - Medical School | Germany | Cancelled |
Thank you for registering! If you will not be able to attend, please contact Tim Beißbarth as soon as possible so that we can notify the people on the waiting list.
How to find us
We meet at the Klinikum Großhadern, Hörsaaltrakt, Floor K O1, Hoersaal I, Marchioninistraße 15, 81377 München.
Großhadern lies in the south-west of Munich (map).
A detailed how-to-get-there is found here (in german only).
For navigating, you might also find the roadmap and the following directions helpful:
• If you arrive in Munich by plane, take the S-bahn S8 ("Airport line") to Marienplatz.
• If you arrive by train, take any S-bahn or U-bahn to Marienplatz.
• From Marienplatz take U6 to final stop Klinikum Großhadern.
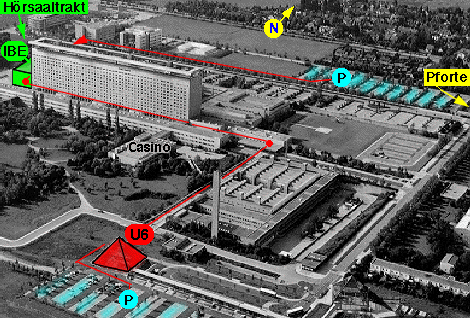
• Leaving the platform, you stand inside a glass pyramid (marked in red in the left figure below).
 |
 |
|
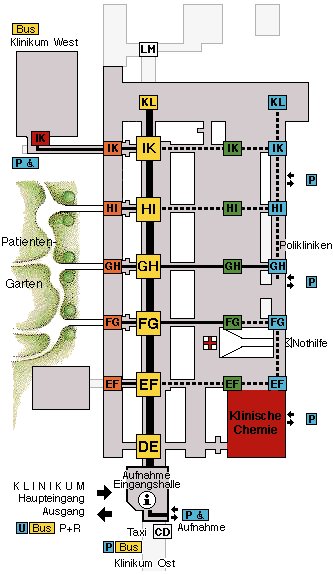
| The red pyramid is the Ubahn-station at which you arrive using U6. The building marked in green is your destination. | This is the inside of the clinical center. The seminarrooms are on the upper right next to the letters "KL". |
Accomodation
You will need to make your own accommodation arrangements. We recommend rooms at the Hotel Neumayr, which lies in Großhadern close to the clinical center. If you want to stay closer to the center of Munich, we recommend hotels in Schwabing, which is 15min by U-bahn from Großhadern. Have a look at Hotel Hauser, Hotel Lettl, or Renner Hotel Savoy.
Further information
Please contact Tim Beissbarth: t.beissbarth@dkfz.de.