Courses in Practical DNA Microarray Analysis 2005
![]()
2005 May 09-12 in Munich
Microarray analysis can be fun! Here are the pictures to prove it.
![]()
| Monday, May 09 - First Analysis steps | ||
| 09.00-09.30 | Introduction and overview | Benedikt Brors |
| 09.30-11.00 | cDNA chips: Quality control and preprocessing | Ulrich Mansmann |
| 11.15-12.15 | Affy Chips: Cel-file versus summary information | Ulrich Mansmann |
| 14.45-17.00 | Exercises: Introduction to R, cDNA data, affy data | Wolfgang Huber |
| Tuesday, May 10 - Exploratory analysis | ||
| 09.00-10.20 | Differential gene expression | Anja von Heydebreck |
| 10.30-11.50 | Clustering | Jörg Rahnenführer |
| 12.00-13.00 | Annotation | Benedikt Brors |
| 14.00-17.00 | Exercises: Differential gene expression Clustering |
v.Heydebreck Rahnenführer,Markowetz |
| Wednesday, May 11 - Molecular Diagnosis | ||
| 09.00-10.20 | Molecular Diagnosis | Rainer Spang |
| 10.30-11.30 | Classification by Nearest Shrunken Centroids and Support Vector Machines |
Florian Markowetz |
| 11.30-12.30 | Model Assessment and Selection | Rainer Spang |
| 13.30-17.00 | Exercises: Molecular Diagnosis | Florian Markowetz |
| Thursday, May 12 - Pathways | ||
| 09.00-10.20 | From a gene list to biological function | Adrian Alexa |
| 10.30-11.30 | Structured Analysis of Microarrays and Differential Coexpression |
Rainer Spang |
| 11.30-12.30 | Scoring pathway activity | Jörg Rahnenführer |
| 13.30-17.00 | Exercises: Group testing, Gene Ontology and time to work on your own data |
Ulrich Mansmann |
Background Knowledge
Ideally, you are interested in mathematical and statistical problems and are familiar with at least one programming language. This course focusses on the practical side of gene expression data analysis. However, data analysis without understanding the statistical background is in general impossible. We strongly recommend you to refresh your mathematical and programming skills before attending the course.
Please use the links to software and literature to prepare yourself before the course begins.
R and Bioconductor
In the afternoon exercises you will learn how to analyze data using the statistical computing environment R [http://www.r-project.org] and BioConductor [http://www.bioconductor.org], an open source software for bioinformatics. R sources and package sources can be downloaded from The Comprehensive R Archive Network at http://cran.r-project.org.
This is a course in microarray analysis -- not an introduction to R. Please read the Introduction to R before the course begins.
Bring your own data
You are encouraged to bring some of your own data to the course (e.g. genepix files or CEL/CDF). We will use this during the exercises. If you expect to have own data only later in the year, it may in fact be advantageous also to register for one of the later courses.
Participants
| • | Adrien Jamain | Imperial College London | |
| • | Anja Weigmann | Institut für Zell- und Molekularpathologie, University of Hannover, Germany | |
| • | Ashraf EL-Sayed | Institute of Animal Breeding Science, Bonn University, Germany | |
| • | Nguyen Trong Ngu | Institute of Animal Breeding Science, Bonn University, Germany. | |
| • | Daniel Kosztyla | TU Berlin, Germany | |
| • | Ulrike Held | Sylvia Lawry Centre for MS Research e.V., Munich, Germany | |
| • | Oliver Thimm | Department of Biology, University of York, UK | |
| • | Thomas Metz | International Rice Research Institute, Los Baños, Laguna, Philippines. | |
| • | Dietmar Fernández Orth | PROGENIKA Biopharma S.A., Derio, Spain | |
| • | David Arteta | Dept. of Pharmacology, University of the Basque Country, Spain | |
| • | Afsaneh Maleki | University of Manchester, UK | |
| • | Manuel Mayhaus | Genopolis Consortium, University of Milano-Bicocca , Italy | |
| • | Ailís Fagan | The National University of Ireland, Dublin, Ireland | |
| • | Ian Jeffery | The National University of Ireland, Dublin, Ireland | |
| • | Walter Glaser | Vienna University Computing Center, Vienna, Austria | |
| • | Maurice Scheer | TFH Berlin, Germany | |
| • | Donald Dunbar | University of Edinburgh, UK | |
| • | Mathias Gebauer | Aventis Pharma Deutschland GmbH, Frankfurt, Germany | |
| • | Stefan Weckx | VIB microarray facility, Leuven, Belgium | |
| • | Sudesh K. Srivastav | Tulane University, New Orleans, US | |
| • | Reinhard Hoffmann | Max-von-Pettenkofer-Institut, Munich, Germany | |
| • | Elena Sarropoulou | Hellenic Centre for Marine Research Crete, Heraklion, Crete, Greece | |
| • | Nadine Zidek | Merck KGaA, Frankfurt, Germany | |
| • | Robert A. Furlong | Department of Pathology, University of Cambridge, UK | |
| • | Sylvia Merk | University of Munich, Germany | |
| • | Dario Greco | University of Helsinki, Finland |
Thank you for registering! This course is fully booked and closed for registration. The course is still open for registration. If you will not be able to attend, please contact Florian Markowetz as soon as possible so that we can notify the people on the waiting list.
How to find us
We meet at the Klinikum Großhadern, Hörsaaltrakt, Floor K O2, Seminarroom 5, Marchioninistraße 15, 81377 München.
Großhadern lies in the south-west of Munich (map).
A detailed how-to-get-there is found here (in german only).
For navigating, you might also find the roadmap and the following directions helpful:
• If you arrive in Munich by plane, take the S-bahn S8 ("Airport line") to Marienplatz.
• If you arrive by train, take any S-bahn or U-bahn to Marienplatz.
• From Marienplatz take U6 to final stop Klinikum Großhadern.
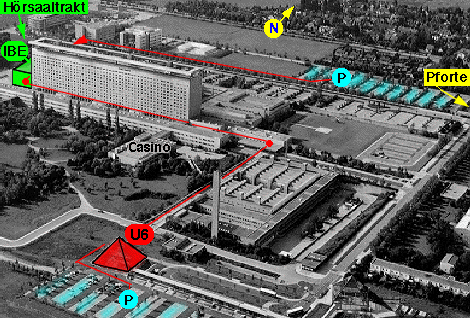
• Leaving the platform, you stand inside a glass pyramid (marked in red in the left figure below).
 |
 |
|
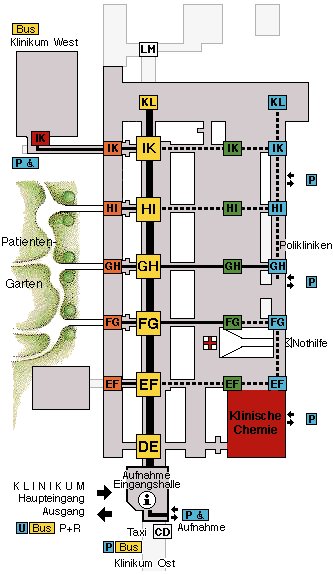
| The red pyramid is the Ubahn-station at which you arrive using U6. The building marked in green is your destination. | This is the inside of the clinical center. The seminarrooms are on the upper right next to the letters "KL". |
Accomodation
You will need to make your own accommodation arrangements. We recommend rooms at the Hotel Neumayr, which lies in Großhadern close to the clinical center. If you want to stay closer to the center of Munich, we recommend hotels in Schwabing, which is 15min by U-bahn from Großhadern. Have a look at Hotel Hauser, Hotel Lettl, or Renner Hotel Savoy.
Further information
Please contact Florian Markowetz: florian.markowetz@molgen.mpg.de.