|
|
Local False Discovery Rate

Stefanie Scheid
Using microarrays it is possible to measure expression of several thousands genes at one time. A high expression value corresponds to an increased abundance of specific mRNA. One important task is to find significant differences between two or more groups of samples. When testing for differential expression simultaneously for all genes, we have to deal with the problem of multiplicity. Declaring positive findings as induced genes, we are interested in the probability that a positive gene is indeed differentially expressed. There exist various kinds of methods to estimate error rates of the top list of differentially expressed genes. One such error rate is the false discovery rate, which is defined as the expected proportion of false positives in the set of positive findings. The error rate is a feature of the list of genes. However, the individual probability of being a false positive is not necessarily the same for all genes in the list.
This project deals with the estimation of the local false discovery rate that measures the probability of single genes being non-induced. In Scheid and Spang (2004a), we proposed a novel estimation procedure, which performed well in comparison to other approaches. The main idea is to use the set of observed p-values and to divide this set into two subsets: one subset ideally follows a uniform distribution and represents the set of non-induced genes. The second subset represents the set of induced genes, for which we do not impose any distributional assumptions except that it does not include a uniform part. The method is implemented in our Bioconductor package twilight. The estimation of the local false discovery rate can be improved by using the Permutation Filtering approach beforehand.
|
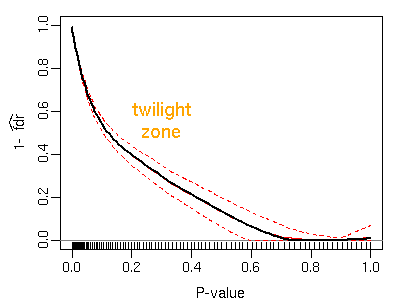
Figure: A typical plot of estimated local FDR: displayed are average estimates with 95% bootstrap confidence intervals. The posterior probability of being induced declines rather slowly and exhibits a broad twilight zone where differential meets non-differential expression.
|

Publications
-
Scheid S and Spang R (2004a):
A stochastic downhill search algorithm for estimating the local false discovery rate
IEEE Transactions on Computational Biology and Bioinformatics 1(3):98-108.
[ abstract | pdf | notice]
-
Scheid S and Spang R (2004b):
Estimation of Local False Discovery Rate - User's Guide to the Bioconductor package TWILIGHT
CompDiag Technical Report Nr. 2004/01 Nov. 2004
[ pdf ]
-
Scheid S and Spang R (2003):
A false discovery rate approach to separate the score distributions of induced and non-induced genes
Proceedings of the 3rd International Workshop on Distributed Statistical Computing.
[ abstract | pdf ]

Software
 |
twilight - Estimating the local false discovery rate
An R/Bioconductor package for two-condition microarray experiments. Includes computation of scores, empirical p-values and local false discovery rates. For further information and download visit the software webpage of twilight. |
|

