|
|
Design of a diagnostic marker panel

Jochen Jäger
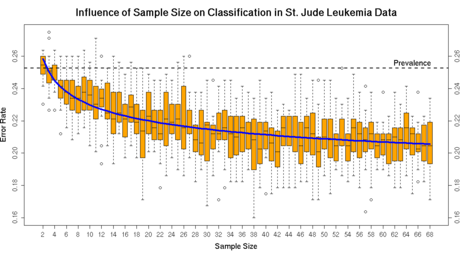
We have developed a novel two-step design for clinical gene expression profiling studies: early marker panel determination (EMPD). In the first step (Phase-1), genome-wide microarrays are used to screen a small number of patients only and to derive a diagnostic marker panel from this data. In the second step (Phase-2), the expression values of only these marker genes are measured in a large group of patients. Phase-2 is used for calibrating the final predictive model and does not require the use of expensive whole genome microarrays, thus making EMPD a cost efficient alternative for current trials.
Analyzing four published clinical microarray datasets we found that in Phase-1 as little as 16 patients per group are sufficient to identify a panel of 10 marker genes. For a marker panel of 100 genes, not more than 10 patients per group are needed. The early decision on the marker panel compromises the final performance of the diagnostic classification only marginally.
The data even suggests that there is an inverse relationship between the number of samples in Phase-1 and the size of the marker panel. As more samples in Phase-1 allow the identification of a more reliable set of markers it is possible to achieve the same relative performance with fewer markers. On the other hand, if it is possible to use many markers, only few samples need to be screened in Phase-1. The results demonstrate that EMPD is a feasible design for cost efficient clinical studies based on gene expression levels. Material, production and handling costs can be saved. Since only few genes in Phase-2 need to be examined, it is possible to utilize small custom diagnostic mRNA arrays or other technologies like qRT-PCR, in-situ hybridization or protein panels. These technologies may also be closer to the clinical phenotype (protein panel) or more precise (qRT-PCR). Since our results in all four studies are consistent other clinical studies as well as ongoing studies may benefit from EMPD.
Future research is concerend with normalization effects of diagnostic chips. Most normalization methods
have strong assumptions that do not hold for small diagnostic microarrays.
When nevertheless using standard normalization methods on these small diagnostic microarrays the effects of differential genes are perturbed so that in the worst case biological effects are lost. One of the assumptions is that most of the genes
are not differentially expressed.
For diagnostic chips, where most of the genes if not all genes are differentially expressed, this assumption does obviously not hold.
Therefore, we propose to include additional normalization genes onto the
small diagnostic microarray. Two strategies are compared:
The first is a data driven univariate selection of normalization genes.
The second is multivariate and based on finding a balanced diagnostic signature.
Our results show that not including additional genes for normalization on small microarrays leads to a loss of diagnostic information.
Using house keeping genes from the literature for normalization
fails to work for certain datasets.
While a data driven selection of additional normalization genes works well,
the best results were obtained using a balanced signature.

Publications
-
Early diagnostic marker panel determination for microarray based clinical studies
Jäger J, Weichenhan D, Ivandic B, Spang R
Statistical Applications in Genetics and Molecular Biology: Vol. 4: No. 1, Article 9.
[ bepress ]
-
Vom Biochip zur maßgeschneiderten Therapie
Jäger J, Spang R
BIOforum 6/2004, pp 50-51
-
Improved Gene Selection for Classification of Microarrays
Jäger
J, Sengupta R, Ruzzo WL
Biocomputing - Proceedings of the 2003 Pacific Symposium, 53-64
[ pdf ]
|


