

|
|
|
Universal prognostic markers
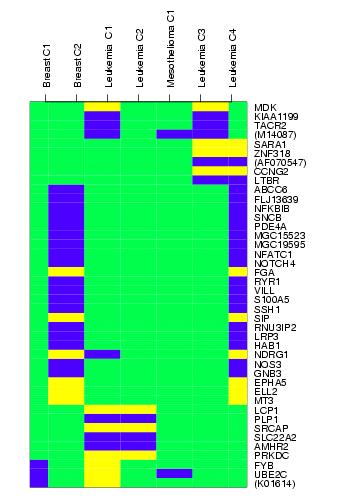
While the majority of oncological microarray studies focus on molecular distinctions in different cancer entities, we focus on detecting common molecular mechanisms in different cancer entities. The method extends previously described concepts by introducing Meta Analysis Pattern Matches. In an analysis of multiple prognostic cancer studies, involving breast cancer, leukemia, and mesothelioma, we are able to identify 42 genes that show consistent up- or down-regulation in patients with a poor disease outcome. Significant up-regulated genes are illustrated as yellow while down-regulated as blue in the right plot. These genes complement the set of previously published candidates for universal prognostic markers in cancer, and suggest the existence of not only specific but also common molecular characteristics, which indicate a poor prognosis in these three types of malignant tumors. The associated common gene list is enriched with genes related to immune response and cell growth and/or cell maintenance. Future interesting research is to detect not only the seperatly independently significant Universal Prognostic Markers , but also those weaker but significant concert ones. Yang and Spang (2005) propose an similarities scores which performs well in identification of those consistent change genes spanning independently multiple studies. The 19 meta-prognostic genes for mesothelioma, prostate and glioma contain a high proportion of know prognostic marker genes and clearly represent biological process involved in tumour progression and formation of metastases. Many of them are extracellular matrix proteins and regulators of its assembly, namely FN1, BGN, POSTN, COL4A1, COL4A2, COL1A2, COL5A2. Most genes have been also identified by other studies of microarraies based on the development of metastases. PTFDS is a novel marker gene, which has not been previously reported. Publications
Software
|
|||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||