Courses in Practical DNA Microarray Analysis 2007
![]()
2007 November 26-29 in Munich
The course is open for registration.
Application: Please send a short note that explains your existing skills and experience with (a) microarrays, (b) statistics, and (c) computer programming and your motivation for taking this course to t.beissbarth@dkfz.de. Additionally, please tell us your NGFN Förderkennziffer (FKZ), if available. You may use the Registration Form.
If you bring your own laptop, be sure to have installed the following packages:
affy, Biobase, marray, multtest, limma, vsn, arrayMagic, cluster, RColorBrewer, hgu95av2, hgu95av2cdf, estrogen,
ALL, annotate, genefilter, multtest, ROC, isis, pamr, e1071, MCRestimate, GlobalAncova, globaltest, GOstats,
hgu133a, Rgraphviz.
Datasets and additional packages used in the practical sessions can be found in the data directory, links to the tutorial slides and practical exercises in the table below.
![]()
![]()
Background Knowledge
Ideally, you are interested in mathematical and statistical problems and are familiar with at least one programming language. This course focusses on the practical side of gene expression data analysis. However, data analysis without understanding the statistical background is in general impossible. We strongly recommend you to refresh your mathematical and programming skills before attending the course.
Please use the links to software and literature to prepare yourself before the course begins.
R and Bioconductor
In the afternoon exercises you will learn how to analyze data using the statistical computing environment R [http://www.r-project.org] and BioConductor [http://www.bioconductor.org], an open source software for bioinformatics. R sources and package sources can be downloaded from The Comprehensive R Archive Network at http://cran.r-project.org.
This is a course in microarray analysis -- not an introduction to R. Please read the Introduction to R before the course begins.
Bring your own data
You are encouraged to bring some of your own data to the course (e.g. genepix files or CEL/CDF). We will use this during the exercises. If you expect to have own data only later in the year, it may in fact be advantageous also to register for one of the later courses.
Participants
| Miguel Angel Pavon Ribas | Inst. Recerca. Hosp. STa. Creu i St. Pau | Spain | |
| Liudmila Liutsko | University of Barcelona, phD student | Spain | Cancelled |
| Simon Geimer | Botanisches Institut in München | Germany | |
| Samuel Arvidsson | University of Potsdam | Germany | |
| Ana Tomasic | Institut Rudjer Boskovic | Croatia | |
| Yannick Le Priol | IMTSSA | France | |
| Sofia Mosci | Universita degli studi di Genova | Italy | |
| Vered Chalifa-Caspi | Ben-Gurion University of the Negev | Israel | Cancelled |
| Raffaella Calligaris | SISSA/ISAS (Internat. School for Advan. Studies) | Italy | |
| Wieslawa Wloszczak | Institute of Bioorganic Chemistry PAS | Poland | Cancelled |
| Subramanian Arun Kumar | University of Reading | United Kingdom. | Cancelled |
| Tan Mei Lan | Universiti Sains Malaysia | Malaysia | |
| Ester Feldmesser | Weizmann Institute of Science | Israel | |
| Ahmet R. Ozturk | Bilkent University / Turkey | Turkey | |
| AKINSOLA LAWAL | YABA COLLEGE TECHNOLOGY | NIGERIA | |
| Peter Konings | Ghent University Hospital | Belgium | |
| Kirstin Meyer | Bayer Schering Pharma | Germany | |
| Arunprababhu Dhanapal | ENEA | Roma, Italy | |
| Divya Mehta | GSF Forschuingszentrum | Germany | |
| Nannan Yang | Faculty of Medicine, Univ. of Tromso, Norway | Norway | Cancelled |
| Peter Novota | University of Goettingen | Germany | Cancelled |
| mohamed adel elsokkary | faculty of pharmacy,Mansoura University | egypt | |
| Aadath Aadath | Aadath | Germany | |
| Mohsen Hajheidari | Max-planck Institute for plant breeding research | Germany | |
| Kutay Karatepe | bilkent university | turkey | Cancelled |
| Emma Cadman | Royal Veterinary College | United Kingdom | |
| Michal Strehovsky | Faculty of Informatics, Masaryk University | Czech republic | |
| mohamed mohamed elsokkary | GSF Neuherberg | Germany | |
| nicolas boulanger | tagc ERM 206 | FRANCE | |
| Xia Dong | TUM | Germany | |
| Damaris Matoke | Kenya Medical Research Institute | Kenya | Waiting List |
| Pieter van Bokhoven | Free University Amsterdam | The Netherlands | |
| Grit Hutter | Klinikum Grosshadern, Med III, KKG Leukemie | Germany | |
| Zlatana Pasalic | GSF-KKG Leukemia | Germany | |
| KAZEEM OLABODE LEMBOYE | NIGERIAN EXPORT IMPORT BANK | LAGOS NIGERIA | Waiting List |
| Md. Asad Ud-Daula | GSF-Institute of Ecological chemistry | D-85764 Neuherberg, Germany | |
| GILBERT OSAYEMWENRE | UNIVERSITY OF BENIN NIGERIA | nigeria | Waiting List |
| Sarrah Ben M | Wageningen university | The Netherlands | |
| Katharina Heim | GSF Forschungszentrum | Germany | |
| necati cakir | University of Trakya, Department of Rheumatology | Turkey | |
| Kivanc Ergen | Kocaeli University, Biophysics Department | Turkey | Waiting List |
| Sukhdeep Singh | Ambala College of Engineering & Applied Research | India | Waiting List |
| Sukhdeep Singh | Ambala College of Engineering & Applied Research | India | Waiting List |
| Ananta paine | Hannover Medical School | Germany | Waiting List |
| Alireza Haghighi | University of Oxford | UK | Waiting List |
| ANANTH RAJAN | CENTRE FOR MARINE SCIENCE AND TECHNOLOGY | INDIA | Waiting List |
| MARY VICTA | BHARTHIAR UNIVERSITY | INDIA | Waiting List |
| Sebastian Waszak | Gene Centre Munich | Germany | Waiting List |
| Benjamin Wasmer | Max-Planck-Institute for Ornithology | Germany | |
| Rafal Woycicki | Warsaw Agricultural University | Poland | |
| Matilda Ulmius | Pure and Applied Biochemistry, Lund University | Sweden | Waiting List |
| Aly Somaa | PAMCHS | Dahrhan 31932. POBox 946 | Waiting List |
| Kristel Van Steen | Ghent University Hospital | Belgium |
Thank you for registering! If you will not be able to attend, please contact Tim Beißbarth as soon as possible so that we can notify the people on the waiting list.
How to find us
We meet at the Klinikum Großhadern, Hörsaaltrakt, Floor K O2, Seminarroom 5, Marchioninistraße 15, 81377 München.
Großhadern lies in the south-west of Munich (map).
A detailed how-to-get-there is found here (in german only).
For navigating, you might also find the roadmap and the following directions helpful:
• If you arrive in Munich by plane, take the S-bahn S8 ("Airport line") to Marienplatz.
• If you arrive by train, take any S-bahn or U-bahn to Marienplatz.
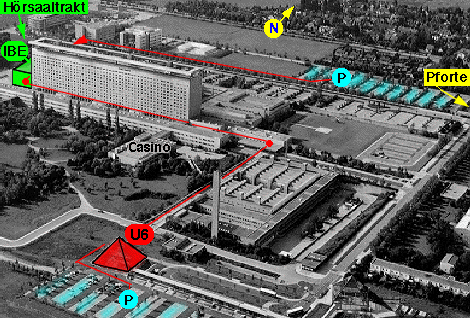
• From Marienplatz take U6 to final stop Klinikum Großhadern.
• Leaving the platform, you stand inside a glass pyramid (marked in red in the left figure below).
 |
 |
|
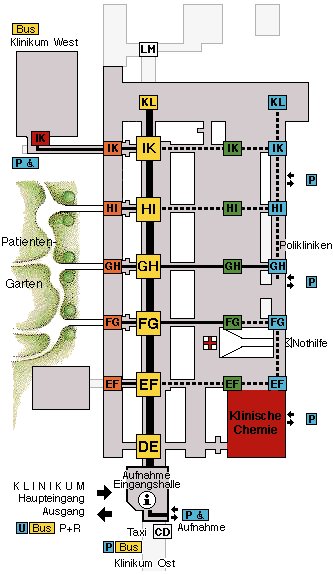
| The red pyramid is the Ubahn-station at which you arrive using U6. The building marked in green is your destination. | This is the inside of the clinical center. The seminarrooms are on the upper right next to the letters "KL". |
Accomodation
You will need to make your own accommodation arrangements. We recommend rooms at the Hotel Neumayr, which lies in Großhadern close to the clinical center. If you want to stay closer to the center of Munich, we recommend hotels in Schwabing, which is 15min by U-bahn from Großhadern. Have a look at Hotel Hauser, Hotel Lettl, or Renner Hotel Savoy.
Further information
Please contact Tim Beissbarth: t.beissbarth@dkfz.de.